Source#
The Source class provides an interface and predefined methods for reading raw
data from file. If source.use_nexus == True the raw data will be saved and
read preferentially from a generated NeXus file, which results in a single
data file with high compression and fast data access.
The Source class provides a dict() of all scans, called scan_dict.
The items in this dictionary are instances of the Scan class, which defines
the common data structure. However, the access to the scan data should NOT
be done via the scan_dict directly, but instead for a deticated scan via
Source.get_scan_data(scan_number) or for a list of scans via
Source.get_scan_list_data(scan_number_list). By this, all features of the
Source class can be utilized.
One key concept of the pyEvalData is the idea, that Scan instances must
only provide the meta information of the scan but not necessarily its data.
By default, the scan data will only be read on request and is then stored in
the Scan object for later use. It is possible, however, to read all data
to each scan directly on first parsing by Source.read_all_data = Ture.
On the other hand, if the data is allocating too much memory, it is possible
to clear the data from the Scan object, directly after accessing it, via
Source.read_and_forget = True. The flag Source.update_before_read enables
parsing the raw source file to search and add new scans before accessing any
other scans. By default the last Scan in the scan_dict will always be
re-created in case new data was added. The flag Source.force_overwrite will
force a full parsing of the raw source file and a complete overwrite of the
NeXus file on every update().
With these set of parameters the user is very flexible in how and if the raw
data is read and stored and can adapt the Source behaviour for different
situations such as on-line anaylsis during a beamtime or post analysis
later at home.
The following examples are meant for introducing the low-level access to raw
data via the Source layer. This can be helpful for direct access to raw data
and integration into existing scripts and applications.
However, user will generally access the Source via the Evaluation class
as described in the upcoming examples.
Setup#
Here we do the necessary import for this example
import matplotlib.pyplot as plt
import numpy as np
import pyEvalData as ped
# define the path for the example data
example_data_path = '../../../example_data/'
SPEC#
A very common data source are SPEC files from the original
Certif spec application as well as from the
open-source alternative Sardana.
The Spec source relies on the great parser provided by
xrayutilities.
spec = ped.io.Spec(file_name='certif_xu.spec',
file_path=example_data_path,
use_nexus=True,
force_overwrite=False,
update_before_read=False,
read_and_forget=True,
nexus_file_path='./')
pyEvalData.io.source - INFO: Update source
pyEvalData.io.source - INFO: parse_raw
pyEvalData.io.source - INFO: Create spec_file from xrayutilities
XU.io.SPECFile.Update: reparsing file for new scans ...
pyEvalData.io.source - INFO: save_all_scans_to_nexus
pyEvalData.io.source - INFO: read_raw_scan_data for scan #32
XU.io.SPECScan.ReadData: scan_32 has been aborted - no data available!
pyEvalData.io.scan - INFO: Scan #32 contains no data!
pyEvalData.io.source - INFO: save_scan_to_nexus for scan #32
pyEvalData.io.source - INFO: read_raw_scan_data for scan #33
XU.io.SPECScan.ReadData: scan_33: 801 27 27
pyEvalData.io.source - INFO: save_scan_to_nexus for scan #33
pyEvalData.io.source - INFO: read_raw_scan_data for scan #34
XU.io.SPECScan.ReadData: scan_34: 801 27 27
pyEvalData.io.source - INFO: save_scan_to_nexus for scan #34
pyEvalData.io.source - INFO: read_raw_scan_data for scan #35
XU.io.SPECScan.ReadData: scan_35: 801 27 27
pyEvalData.io.source - INFO: save_scan_to_nexus for scan #35
pyEvalData.io.source - INFO: read_raw_scan_data for scan #36
XU.io.SPECScan.ReadData: scan_36: 401 27 27
pyEvalData.io.source - INFO: save_scan_to_nexus for scan #36
As described above, the scan data and meta information should be accessed via
spec.get_scan_data(scan_number).
Here, data is returned as numpy.recarray and meta is a dict().
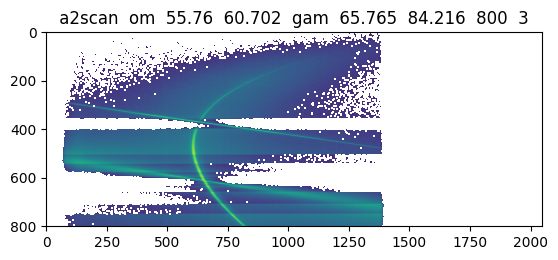
data, meta = spec.get_scan_data(33)
plt.figure()
plt.imshow(np.log10(data['MCA']))
plt.title(meta['cmd'])
plt.show()
One can also directly work with the Scan objects, which provides nearly the
same functionality as above but misses to automatically read_and_forget the
data. Below, the example from above is reproduces with the Scan. Here one can
also easily access the meta information as attributes of the Scan instance.
scan = spec.get_scan(33)
plt.figure()
plt.imshow(np.log10(scan.MCA))
plt.title(scan.cmd)
plt.show()

It is also possible to access the scan directly as attribute of the source which is fully aquivalent to the last example.
plt.figure()
plt.imshow(np.log10(spec.scan33.MCA))
plt.title(spec.scan33.cmd)
plt.show()

Sardana NeXus#
The NeXus file format is a common standard in
science for storing and exchanging data. It is based on
hdf5 files with a hierarchical
structure. As an example we read the NeXus files as created by Sardana.
As we can directly access the data from the NeXus files, there is no need to
enable additional NeXus export so one can set source.use_nexus = False.
sarnxs = ped.io.SardanaNeXus(file_name='sardana_nexus.h5',
file_path=example_data_path,
use_nexus=False,
force_overwrite=False,
update_before_read=False,
read_and_forget=True)
pyEvalData.io.source - INFO: Update source
pyEvalData.io.source - INFO: parse_raw
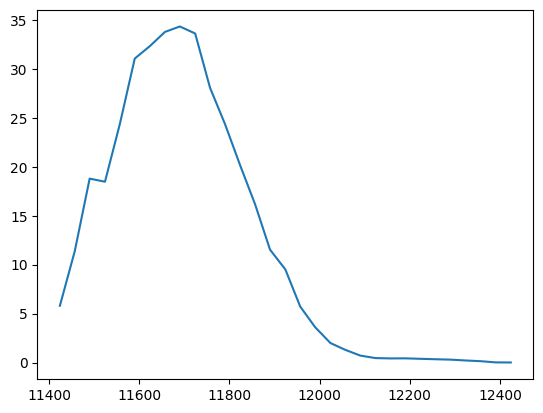
plt.figure()
plt.plot(sarnxs.scan435.mhor, sarnxs.scan435.pmPump)
plt.show()
pyEvalData.io.source - INFO: read_raw_scan_data for scan #435
hdf5 - PAL FEL#
hdf5 is also a very common data
format. In this example the raw data was measured at PAL FEL in South Korea.
The individual files are grouped in a single folder per scan. This is handled
by the fprint-statement for the file_name is is automatically evaluated
to provide the correct file and folder name for a given scan number.
pal = ped.io.PalH5(name='2020_12_Schick',
file_name='{0:07d}',
file_path=example_data_path+'/pal_fel',
use_nexus=False,
force_overwrite=False,
update_before_read=True,
read_and_forget=True,
nexus_file_path='./',
nexus_file_name='2020_12_Schick')
pyEvalData.io.source - INFO: Update source
pyEvalData.io.source - INFO: parse_raw
Again we have easy access to the raw data similar to the former examples.
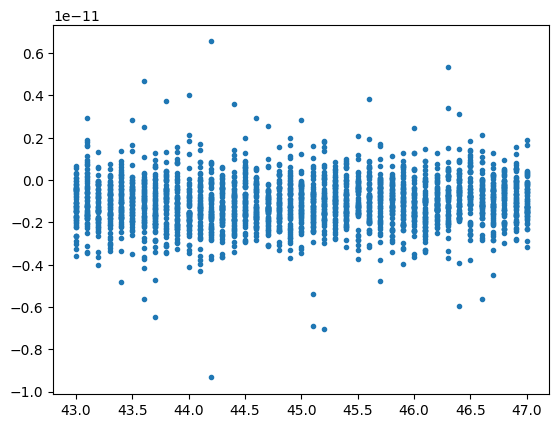
plt.figure()
plt.plot(pal.scan9.th, pal.scan9.digi1_1, '.')
plt.show()
pyEvalData.io.source - INFO: read_raw_scan_data for scan #9
Composite Sources#
idea for a future release
One specific idea of the Source class is to provide also composite models of
pre-defined Source classes. An example is the Spec source for reading SPEC
data files and a Source class to read camera images from a folder structure,
that have been acquired simultaneously with the SPEC file.
This would require to specify the two independent Souce objects and then add
them to a single CompositeSource, e.g. by:
spec = ped.io.Spec(file_name, file_path)
ccd = ped.io.Pilatus(image_pattern, image_base_path)
comp_source = spec + ccd
After this step the workflow is the same as with all other Source classes.
The concatenation should also allow for multiple items. Special care must be
take for duplicate motor and counter names. This would require a prefix
scheme similar to the lmfit package.